| Region plot | Region plot detail | Dotplot |
 |  |  |
| [jpg] [pdf] | [jpg] [pdf] | [jpg] [pdf] |
| CRM: | PCE7002 | |
|---|---|---|
| Overlaps: | eve stripes 3/7 | |
| Reference: | Harding/Goto 1989 | |
| Berman 2002 ID: | ||
| Crm Activity: | + | |
| 5' Primer: | ||
| 3' Primer: | ||
| D.mel fasta: | dmel unflanked | |
| pCRM region: | genomic region | |
| Chrom arm: | 2R | |
| pCRM start: | 5,035,494 | |
| pCRM end: | 5,036,771 | |
| pCRM len: | 1,278 | |
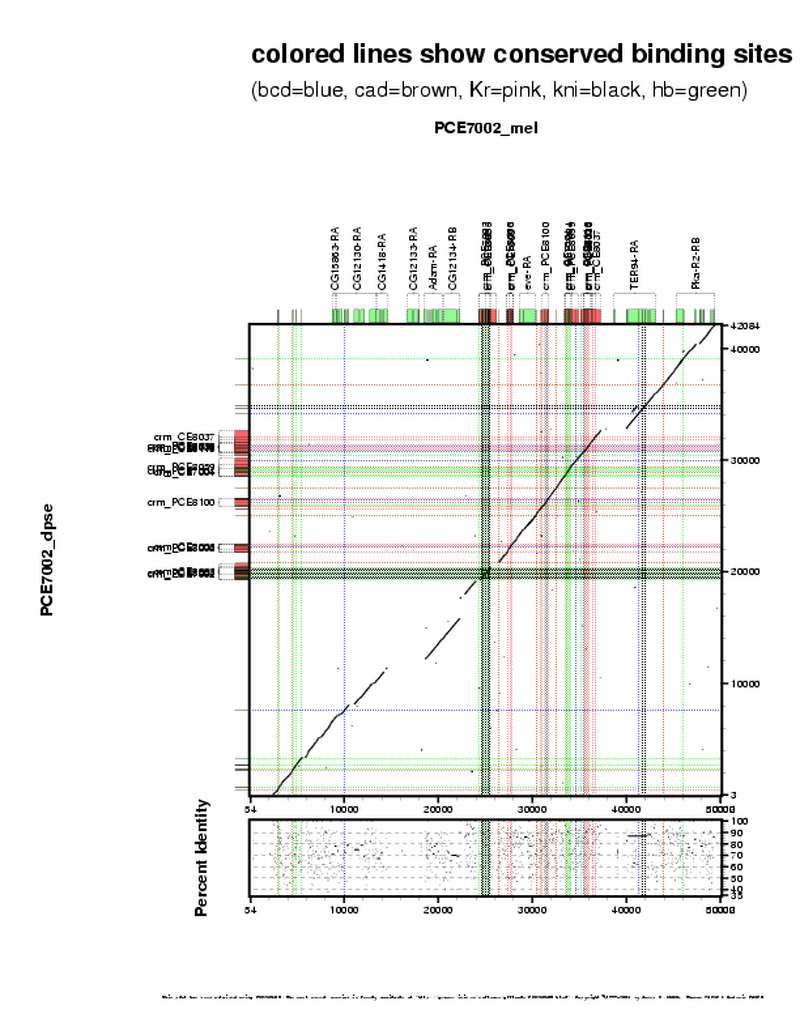
| D.pse region homologous: | 1 | |
| D.pse fasta: | dpse unflanked | |
| dpse arm: | Dpse_3213009_3212129_1_42085 | |
| dpse start: | 19,302 | |
| dpse end: | 20,415 | |
| dpse len: | 1,114 | |
| Flanked D.mel fasta: | dmel flanked | |
| Flanked D.pse fasta: | dpse flanked | |
| LAGAN alignment (readable): | alignment | |
| LAGAN alignment (mfa): | alignment | |
| D.mel binding site xml: | xml | |
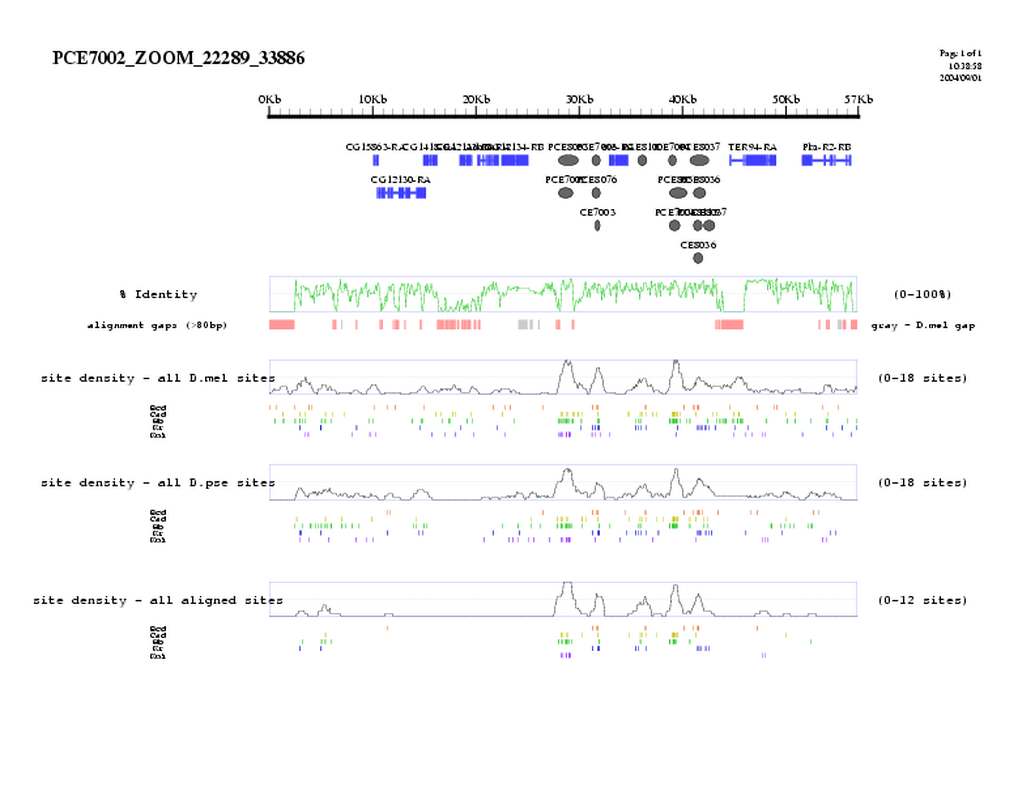
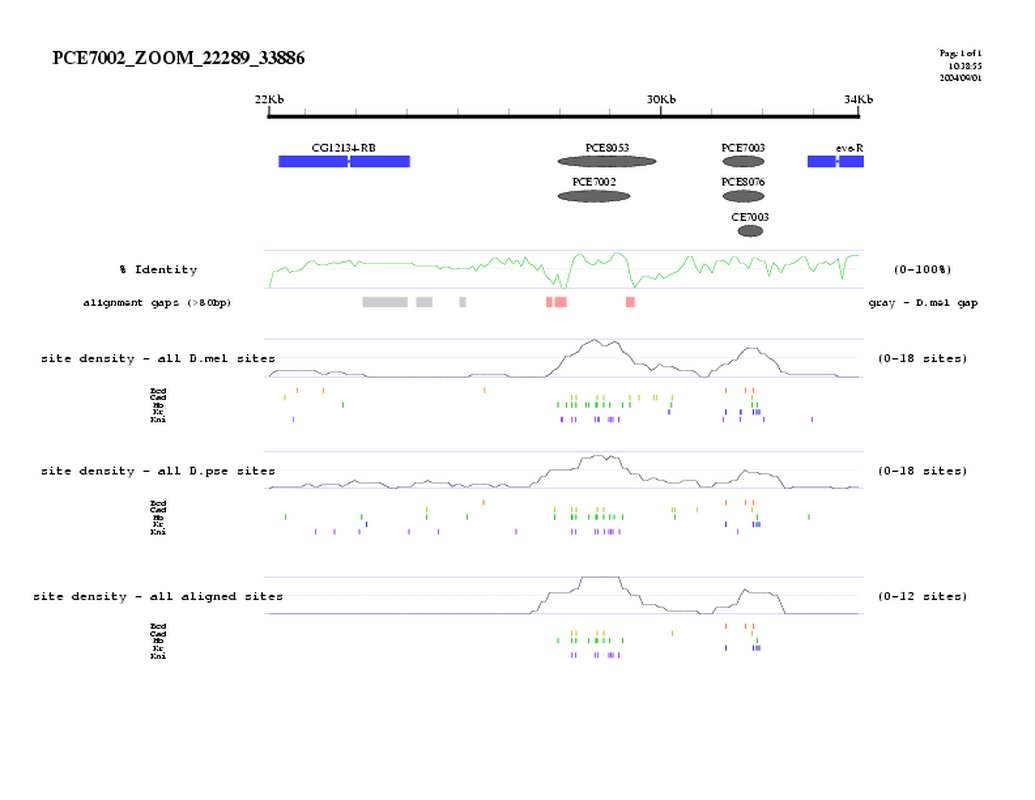
| overall site conservation z-score: | 13.6 | |
| Percent identity: | 60.6% | |
| D.mel sites: | 28 | |
| D.pse sites: | 25 | |
| Aligned sites: | 21 | |
| Aligned + preserved sites: | 25 | |
| D.mel site dens: | 21.9 | |
| D.pse site dens: | 22.4 | |
| Aligned site dens: | 16.4 | |
| Aligned + preserved site dens: | 19.6 | |
| pCRM position relative to 5' gene: | downstream | |
| pCRM position 5' gene relative: | 3,713 | |
| 5' gene CG: | CG12134 | |
| 5' gene: | CG12134 | |
| 5' gene insitu: | CG12134 insitu | |
| 5' gene flybase: | CG12134 flybase | |
| 5' gene start: | 5,031,781 | |
| 5' gene end: | 5,033,416 | |
| 5' gene txl start: | 5,032,291 | |
| pCRM position 3' gene relative: | upstream | |
| pCRM position relative to 3' gene: | -2,952 | |
| 3' gene CG: | CG2328 | |
| 3' gene: | eve | |
| 3' gene insitu: | eve insitu | |
| 3' gene flybase: | eve flybase | |
| 3' gene start: | 5,039,723 | |
| 3' gene end: | 5,041,412 | |
| 3' gene txl start: | 5,040,047 | |