pCRM PCE8067
| CRM: |
| PCE8067 |
|---|
| Overlaps: |
| ftz upstream (partial) |
|---|
| Reference: |
| Hiromi, 1985 |
|---|
|
| D.mel fasta: |
| dmel unflanked |
|---|
| pCRM region: |
| genomic region |
|---|
| Chrom arm: |
| 3R.3 |
|---|
| pCRM start: |
| 2,682,314 |
|---|
| pCRM end: |
| 2,684,591 |
|---|
| pCRM len: |
| 2,278 |
|---|
|
| D.pse fasta: |
| dpse unflanked |
|---|
| dpse arm: |
| 3,212,942 |
|---|
| dpse start: |
| 1,780,248 |
|---|
| dpse end: |
| 1,782,485 |
|---|
| dpse len: |
| 2,238 |
|---|
|
| Flanked D.mel fasta: |
| dmel flanked |
|---|
| Flanked D.pse fasta: |
| dpse flanked |
|---|
| LAGAN alignment (readable): |
| alignment |
|---|
| LAGAN alignment (mfa): |
| alignment |
|---|
|
| D.mel binding site xml: |
| xml |
|---|
|
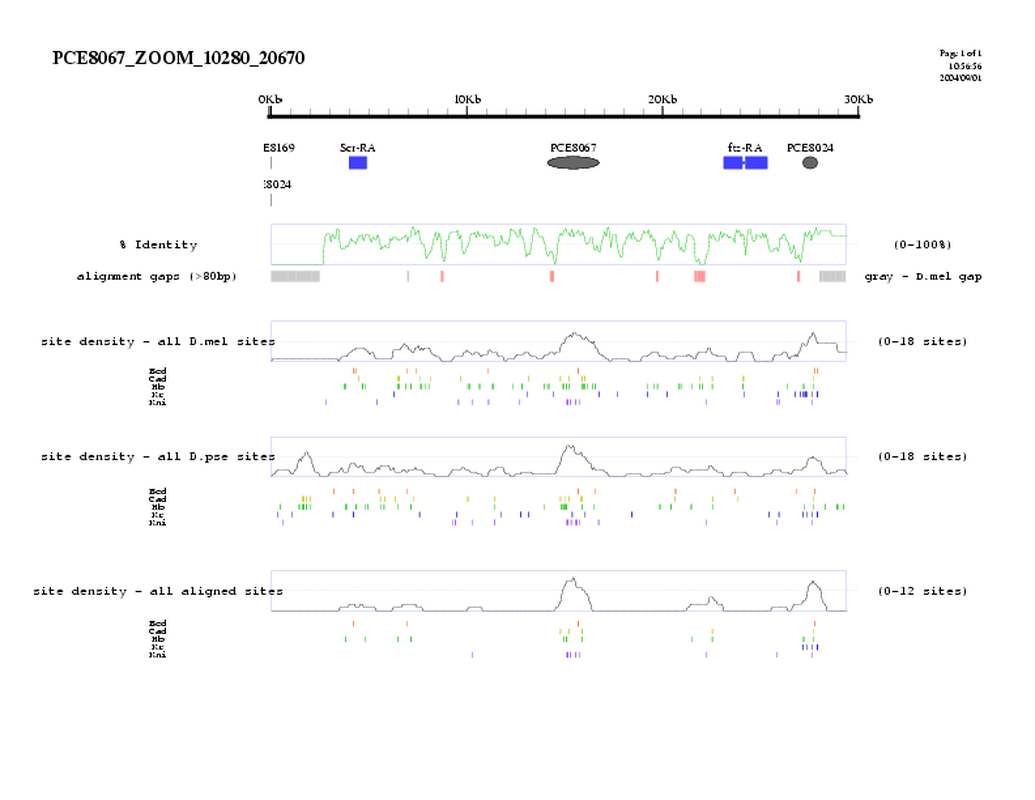
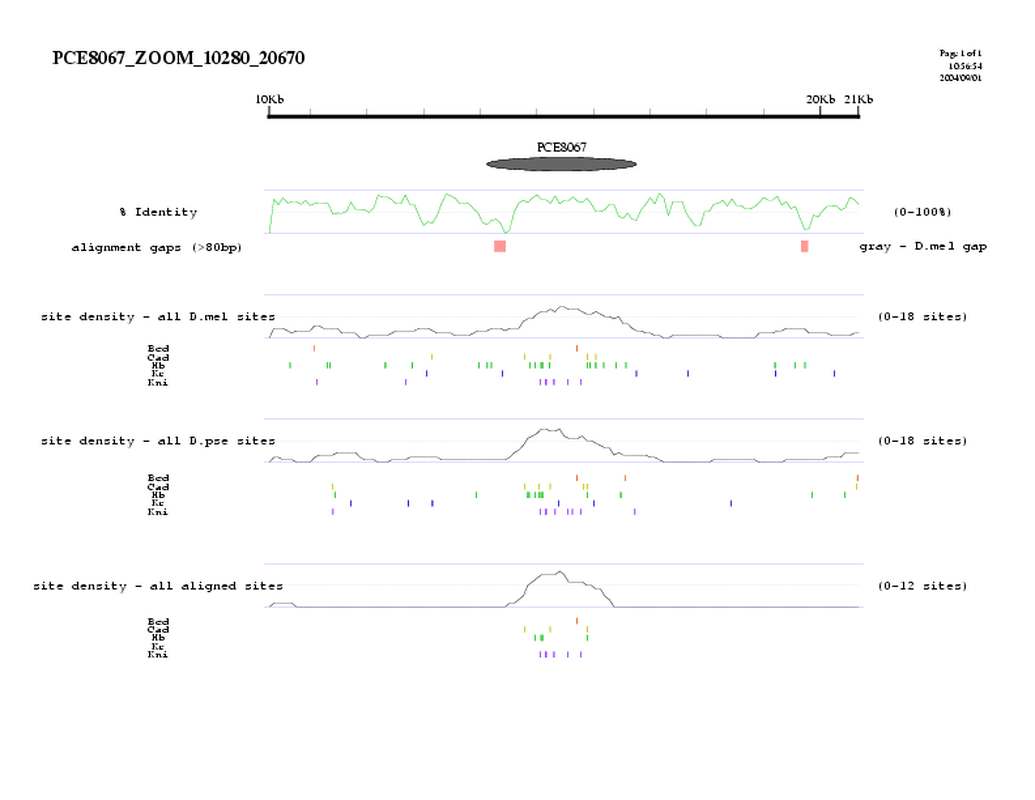
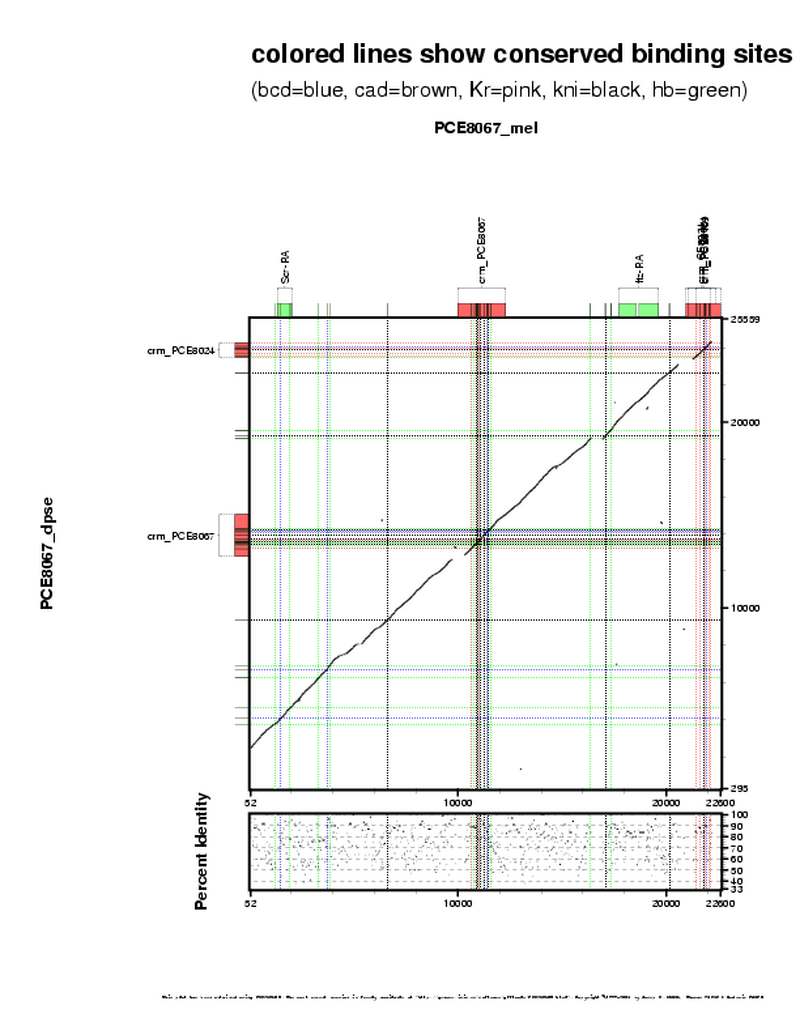
| overall site conservation z-score: |
| 8.3 |
|---|
|
| percent iden: |
| 58.0% |
|---|
|
| D.mel sites: |
| 27 |
|---|
| D.pse sites: |
| 26 |
|---|
| Aligned sites: |
| 15 |
|---|
| Aligned + preserved sites: |
| 22 |
|---|
|
| D.mel site dens: |
| 11.9 |
|---|
| D.pse site dens: |
| 11.6 |
|---|
| Aligned site dens: |
| 6.6 |
|---|
| Aligned + preserved site dens: |
| 9.7 |
|---|
|
| pCRM position relative to 5' gene: |
| upstream |
|---|
| pCRM position 5' gene relative: |
| -7,972 |
|---|
| 5' gene CG: |
| CG1030 |
|---|
| 5' gene: |
| Scr |
|---|
| 5' Gap/pair-rule annotation: |
| FlyBase |
|---|
| 5' gene insitu: |
| |
|---|
| 5' gene flybase: |
| Scr flybase |
|---|
| 5' gene start: |
| 2,674,342 |
|---|
| 5' gene end: |
| 2,648,842 |
|---|
| 5' gene txl start: |
| 2,667,903 |
|---|
|
| pCRM position 3' gene relative: |
| -5,455 |
|---|
| pCRM position relative to 3' gene: |
| upstream |
|---|
| 3' gene CG: |
| CG2047 |
|---|
| 3' gene: |
| ftz |
|---|
| 3' Gap/pair-rule annotation: |
| FlyBase |
|---|
| 3' gene insitu: |
| |
|---|
| 3' gene flybase: |
| ftz flybase |
|---|
| 3' gene start: |
| 2,690,046 |
|---|
| 3' gene end: |
| 2,691,951 |
|---|
| 3' gene txl start: |
| 2,690,116 |
|---|
|
| Additional Gap/pair-rule gene within 20kb: |
| |
|---|
| Gap/pair-rule annotation: |
| |
|---|
| pCRM relative position: |
| |
|---|