pCRM PCE8825
| CRM: |
| PCE8825 |
|---|
| Overlaps: |
| 0 |
|---|
| Reference: |
| 0 |
|---|
|
| D.mel fasta: |
| dmel unflanked |
|---|
| pCRM region: |
| genomic region |
|---|
| Chrom arm: |
| 2R.3 |
|---|
| pCRM start: |
| 10,684,893 |
|---|
| pCRM end: |
| 10,685,730 |
|---|
| pCRM len: |
| 838 |
|---|
|
| D.pse fasta: |
| dpse unflanked |
|---|
| dpse arm: |
| 3,212,958 |
|---|
| dpse start: |
| 92,663 |
|---|
| dpse end: |
| 93,638 |
|---|
| dpse len: |
| 976 |
|---|
|
| Flanked D.mel fasta: |
| dmel flanked |
|---|
| Flanked D.pse fasta: |
| dpse flanked |
|---|
| LAGAN alignment (readable): |
| alignment |
|---|
| LAGAN alignment (mfa): |
| alignment |
|---|
|
| D.mel binding site xml: |
| xml |
|---|
|
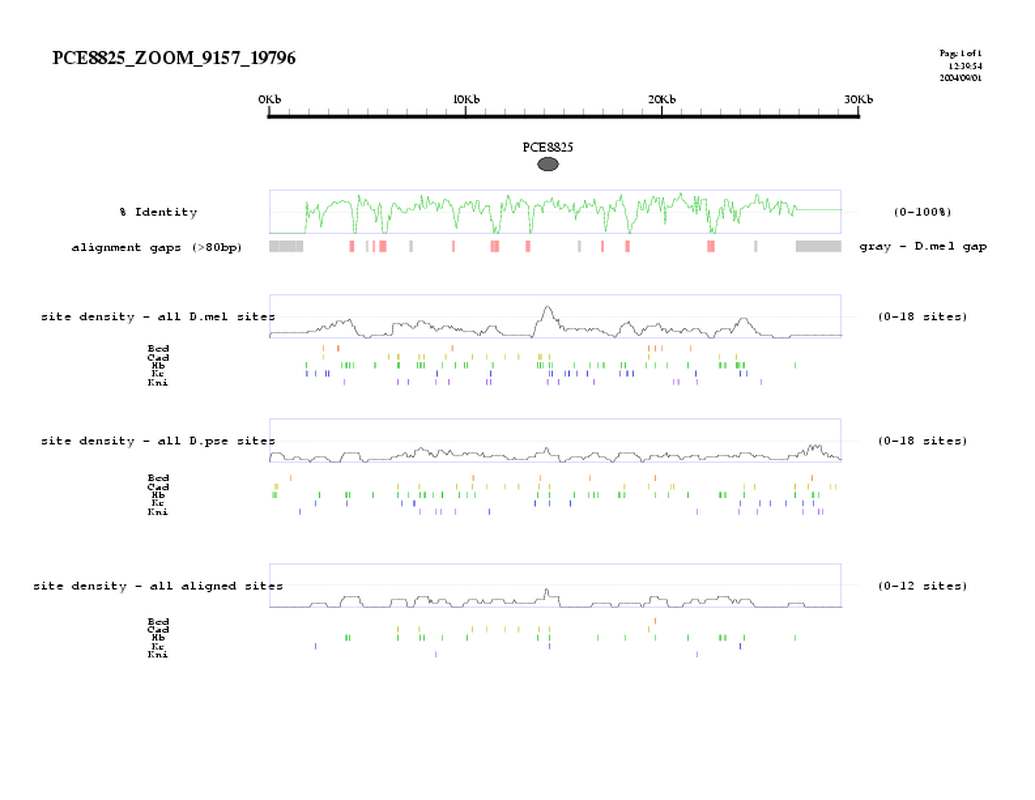
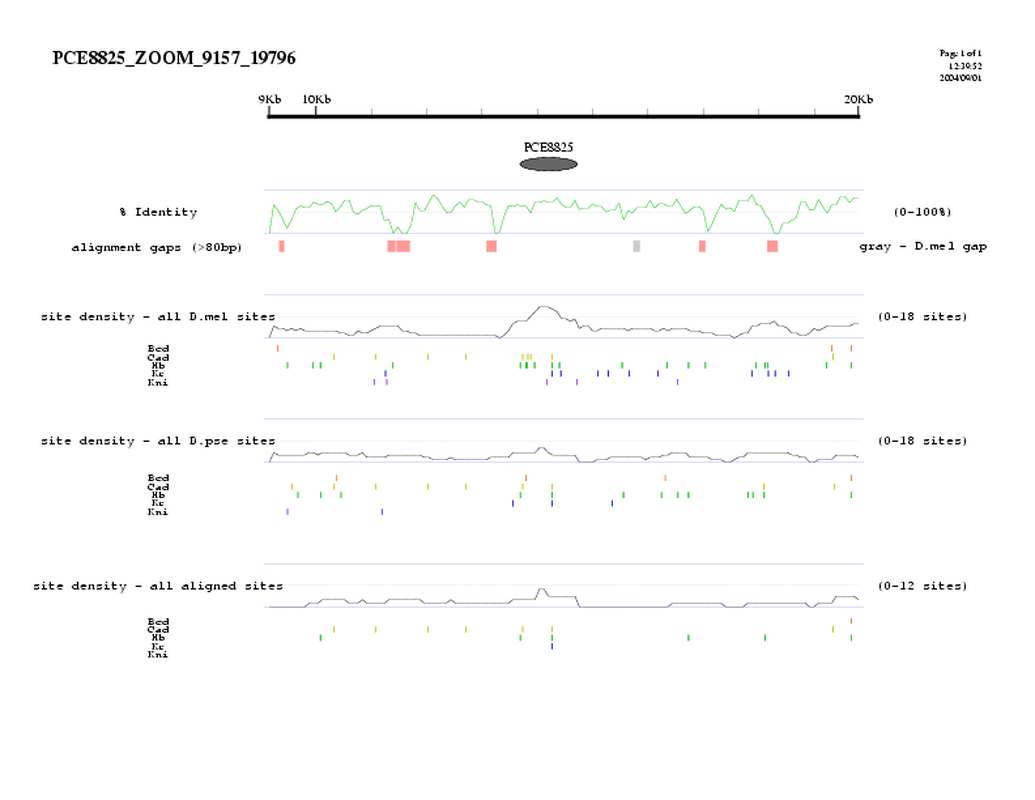
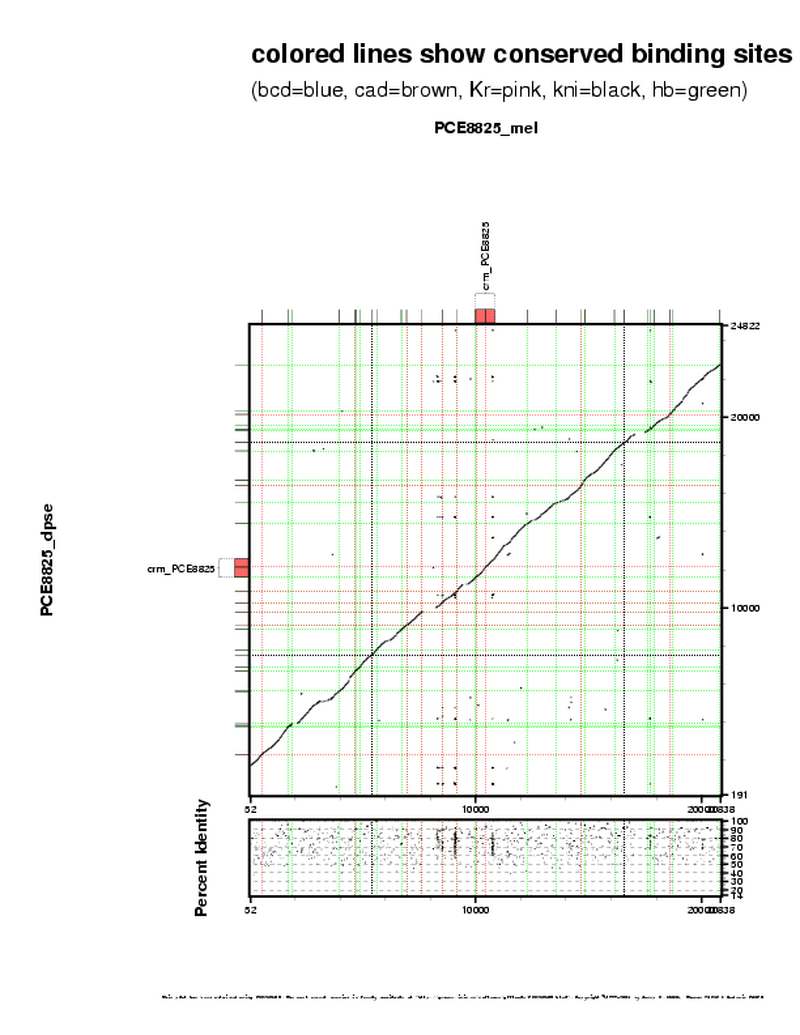
| overall site conservation z-score: |
| 3.0 |
|---|
|
| percent iden: |
| 69.0% |
|---|
|
| D.mel sites: |
| 14 |
|---|
| D.pse sites: |
| 6 |
|---|
| Aligned sites: |
| 5 |
|---|
| Aligned + preserved sites: |
| 5 |
|---|
|
| D.mel site dens: |
| 16.7 |
|---|
| D.pse site dens: |
| 6.1 |
|---|
| Aligned site dens: |
| 6.0 |
|---|
| Aligned + preserved site dens: |
| 6.0 |
|---|
|
| pCRM position relative to 5' gene: |
| downstream |
|---|
| pCRM position 5' gene relative: |
| +14902 |
|---|
| 5' gene CG: |
| CG30465 |
|---|
| 5' gene: |
| CG30465 |
|---|
| 5' Gap/pair-rule annotation: |
| |
|---|
| 5' gene insitu: |
| |
|---|
| 5' gene flybase: |
| CG30465 flybase |
|---|
| 5' gene start: |
| 10,669,991 |
|---|
| 5' gene end: |
| 10,670,613 |
|---|
| 5' gene txl start: |
| 10,670,097 |
|---|
|
| pCRM position 3' gene relative: |
| +43063 |
|---|
| pCRM position relative to 3' gene: |
| downstream |
|---|
| 3' gene CG: |
| CG8205 |
|---|
| 3' gene: |
| fus |
|---|
| 3' Gap/pair-rule annotation: |
| |
|---|
| 3' gene insitu: |
| |
|---|
| 3' gene flybase: |
| fus flybase |
|---|
| 3' gene start: |
| 10,728,793 |
|---|
| 3' gene end: |
| 10,723,740 |
|---|
| 3' gene txl start: |
| 10,727,829 |
|---|
|
| Additional Gap/pair-rule gene within 20kb: |
| |
|---|
| Gap/pair-rule annotation: |
| |
|---|
| pCRM relative position: |
| |
|---|