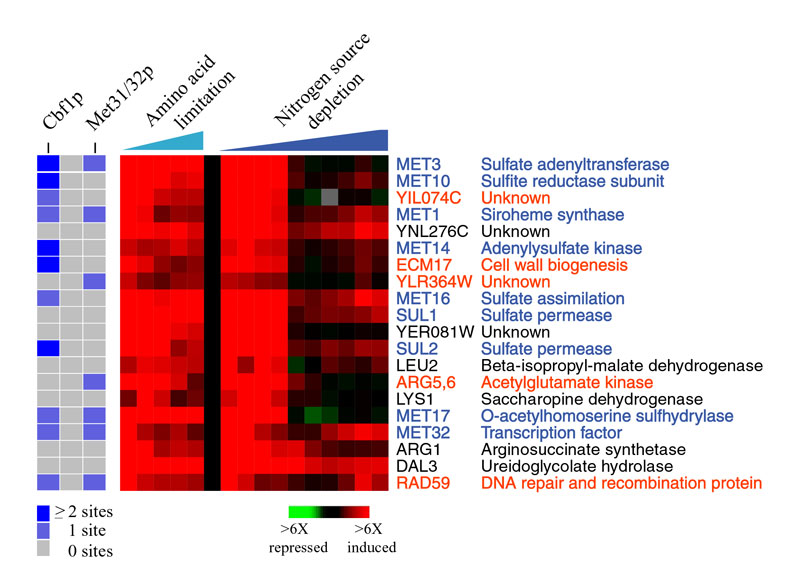
The expression of the top-20 genes in cluster 2 are shown (right panel) along with the copy number of the Cbf1p and Met31/32p binding sites within 800bp upstream of each gene (left panel). Genes that encode proteins known to participate in sulfur fixation and methionine synthesis are shown in blue. Gene that contain either the Cbf1p and/or Met31/32p binding sites and belong significantly to this cluster are labeled in orange. These genes are likely to be coregulated with the methionine biosynthesis genes and may serve a similar cellular role.
For example, Rad59p has been extensively studied for its role in homologous recombination and DNA damage repair. The RAD59 gene is not only highly correlated in expression to the methionine biosynthesis genes, but it contains both the Cbf1p and Met31/32p promoter elements that are known to govern the expression of the methionine genes. The probability of these observations occurring by chance is 10-6, strongly suggesting that RAD59 is coregulated with genes involved in methionine synthesis. Why RAD59 would be coregulated with these genes is unclear but may be related to the alternative role of the transcription factor Cbf1p in centromere function and chromosome segregation; this putative link is supported by the fact that the cell-cycle dependent expression of the methionine biosynthesis genes peaks in mitosis when chromosomes are segregated (Spellman et al. 1998).