Transcriptional Enhancers
In 1983 the labs of Walter Schaffner and Susumu Tonegawa announced the discovery of a class of sequences in animal genomes that regulate gene expression at a distance, sequences we now know as transcriptional "enhancers". We now know that there are tens of thousands of enhancers in animal genomes, and that they play a critical role in the generation of spatial and temporal patterns of gene expression during development. However, despite over thirty years of extensive, we still have a poor understanding of how enhancers work.
We use a combination of experimental, evolutionary and computational genomics, high-resolution imaging, genome engineering and classical biochemistry and genetics to study the ~1,000 enhancers active in the early fly embryo.
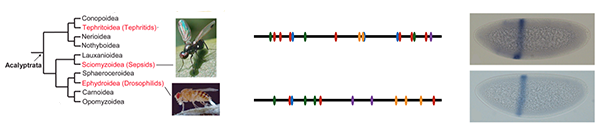
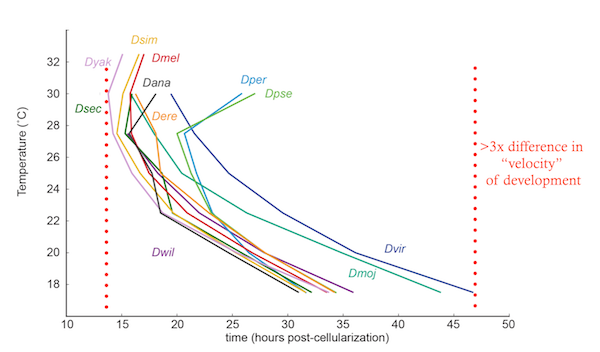
Our current focus is understanding how enhancer identity is established during early embryogenesis.
Lab publications on enhancers
Lusk RW, Eisen MB (2010). Evolutionary mirages: selection on binding site composition creates the illusion of conserved grammars in Drosophila enhancers.
Journal PubMed PMC
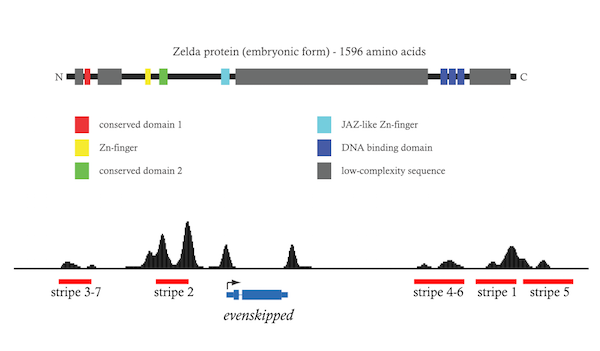
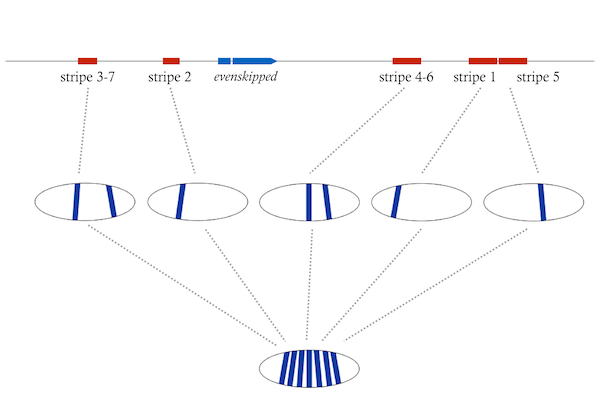
Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB (2011). Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition.
Journal PubMed PMC
Li XY, Harrison MM, Villalta JE, Kaplan T, Eisen MB (2014). Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition.
Journal PubMed PMC Preprint Data:GEO