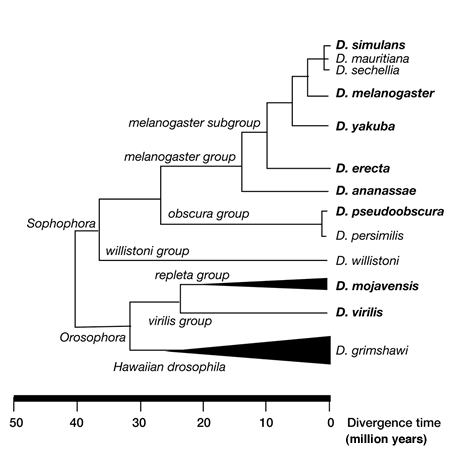
Several groups have produced alignments of various subsets of these Drosophila species. Please remember that accurate multiple-species alignment is a challenging problem, and these alignments must be viewed as works in progress.
Colin Dewey in Lior Pachter's group at UC Berkeley has developed software - called MERCATOR to perform multiple whole-genome orthology map construction, which they have combined with their MAVID multiple aligner to produce multiple alignments. Raw alignments of several Drosophila species, as well as a multi-species browser for each alignment, are available here.
Aaron Darling in Nicole Perna's group at UW-Madison has developed multiple-alignment software called MAUVE. The alignments and viewing software are available here.
Dan Pollard and Venky Iyer in Michael Eisen's group at UC Berkeley/LBNL have produced alignments using MLAGAN to align orthologous blocks identified using orthologous coding genes, syteny and nucleotide similarity. These alignments are available here.
|